RADICaL (Zhu et al. 2022)
RADICaL is an extension of AutoLFADS for application to 2-photon (2p) calcium imaging data. AutoLFADS is the combination of Latent Factor Analysis via Dynamical Systems (LFADS), a deep learning method to infer neural population dynamics, with Population-Based Training (PBT), an automatic hyperparameter tuning framework as described in this paper. RADICaL incorporates two major innovations over AutoLFADS (see paper link). First, RADICaL’s observation model was modified to better account for the statistics of deconvolved calcium events. Second, RADICaL integrated a novel neural network training strategy, selective backpropagation through time (SBTT), that exploits the staggered timing of 2p sampling of neuronal populations to recover network dynamics with high temporal precision.
RADICaL takes in deconvolved calcium events as input, and outputs inferred underlying event rates. The inferred event rates can then be mapped to the ground truth Lorenz states to evaluate how effective RADICaL recovers the latent states.
Args
-Input: (h5 file) Input data format is {num_neurons, num_samples, num_trials}. The mat file can be converted to h5 file using the convert_h5.m code provided in the scripts folder in the demo link. The validation ratio can be set in the code for dividing the dataset into training and validation sets. Make sure you name the dataset with initials the same as the “run_name” in the config file. For example, if you choose the run_name as run001 then you can name the h5 file as run001_data.h5.
-Config: (yaml) A YAML file containing parameters and hyperparameters values for training the model. Use the config file template in the demo link.
Outputs
The results will be saved in the S3 bucket inside the results folder. It contains two folders-
-pbt_best_model: This folder stores the output h5 files which contain the inferred rate parameters for train and valid sets. The post-analysis script will take in these h5 files to compute inferred event rates and more advanced analyses will be performed based on the inferred event rates.
-run_name: This folder stores the detailed training logs, searched hyperparameters, and model checkpoints for each individual PBT worker of the last generation of training. For details in PBT training, please refer to this paper and AutoLFADS google cloud tutorial.
Example Dataset
We have provided a synthetic 2-photon calcium imaging dataset in the demo link for reference. This sample dataset contains simulated deconvolved calcium events that are derived from a Lorenz system. This synthetic dataset is ideal to test out RADICaL as it is intrinsically low-dimensional, and the event rates inferred by RADICaL can easily be mapped to the ground truth latent states to evaluate the effectiveness of RADICaL.
Briefly, spike trains are generated by simulating a population of neurons whose firing rates are linked to the state of the Lorenz system. Realistic fluorescence traces are generated from the spike trains and deconvolved to extract calcium events. Importantly, the sampling times for different neurons are staggered to simulate 2p laser scanning sampling times.
First, open the map_to_latents.m script in the demo link. Fill in the folder path to the scripts directory and RADICaL output directory.
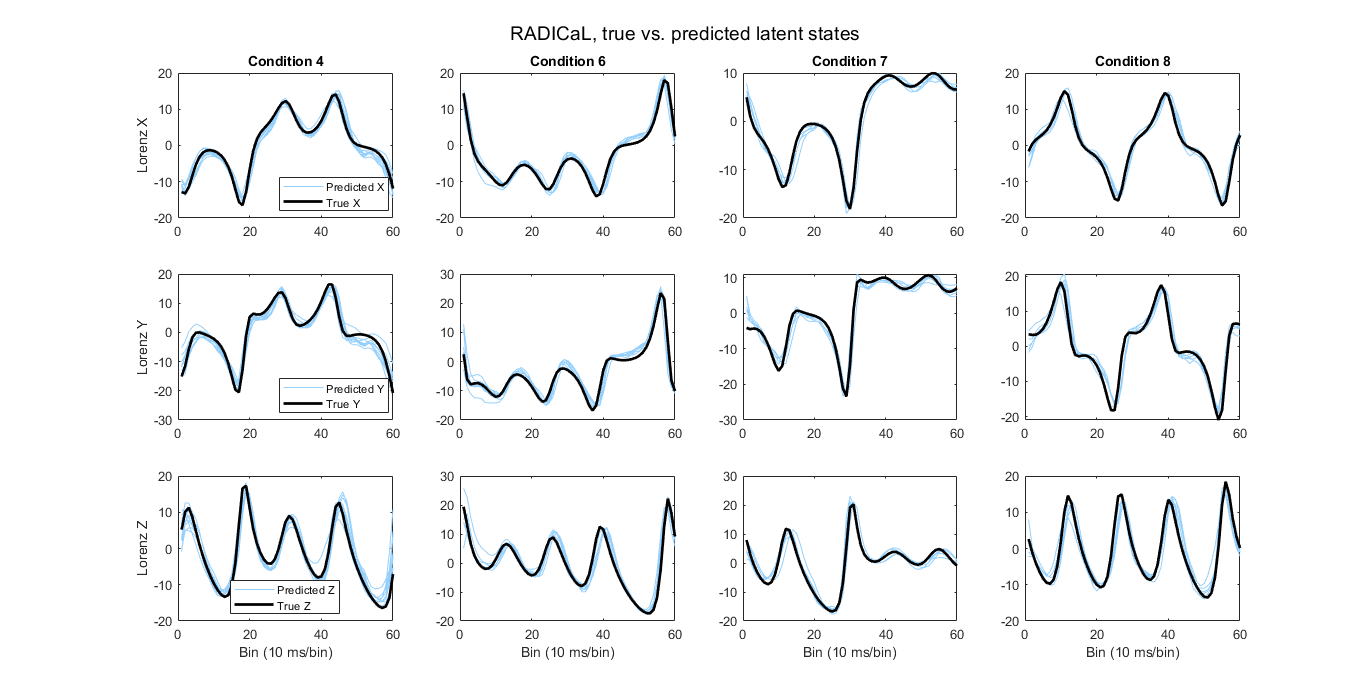
Then, you can run the script. RADICaL inferred rates are mapped to the ground truth Lorenz states by training a ridge regression. The script will then generate R^2 (coefficient of determination) value, which represents how well the predicted latent states match to the true latent states for the testing trials, as well as plot the predicted latent states of several example conditions against their ground truth latent states as shown below.
Hyperparameters
-Frequently Changed Parameters:
| Name | Description |
|---|---|
| run_name | prefix of input dataset h5 file and name of the output folder containing results |
| epochs_per_generation | Number of epochs per each generation |
| max_generations | Maximum number of generations. It may actually take lesser generations to converge to the best model, depending on the converging criterion defined in 'num_no_best_to_stop' and 'min_change_to_stop' |
| num_no_best_to_stop | If no improvement is seen in the best worker for these many successive generations, PBT is terminated even before the max_generations are completed |
| percent_change_to_stop | If the improvement in the best worker over successive generations is less than this quantity, the improvement is considered to be 0. Default value is 0.00. If the num_no_best_to_stop is set to 5 and percent_change_to_stop is set to 0.05 , the PBT training will terminate if for 5 successive generations the improvement in the best worker is less than 0.05% over the previous best worker |
| max_processes | Max_processes are the total number of processes running simultaneously in the instance. Max_processes is usually set to be a multiple of the number of GPUs. Currently we are using the p2.8xlarge EC2 instance with 8 GPUs. Hence max_processes should be 8 or a multiple of 8 depending upon the number of processes to be run on each GPU. For example, if the user wants to run two processes per GPU, then max_processes can be set as 16. |
| num_workers | The number of workers running for each generation. Each worker runs a single model. |
-Explorable Parameters:
| Name | Description |
|---|---|
| learning_rate_init | learning rate initialisation parameter |
| keep_prob | Dropout keep probability |
| keep_ratio | Coordinated dropout input keep probability |
| l2_gen_scale | L2 regularization cost for the generator |
| l2_con_scale | L2 cost for the controller |
| kl_co_weight | Strength of KL weight on controller output KL penalty |
| kl_ic_weight | Strength of KL weight on initial conditions KL penalty |
| gamma_prior | PBT-searchable prior for the scaling factors of sigmoid transformation of ZIG parameters |
| l2_gamma_distance_scale | Penalty applied to the difference between trainable scaling factors and priors for the sigmoid transformation |
-Fixed Parameters:
| Name | Description |
|---|---|
| batch_size | Batch_size to use during training |
| valid_batch_size | Batch_size to use during validation |
| factors_dim | Number of factors from the generator |
| ic_dim | Dimension of hidden state of the initial condition encoder |
| ic_enc_dim | Dimension of hidden state of the initial condition encoder |
| gen_dim | Size of hidden state for generator |
| co_dim | Dimensionality of the inferred inputs by the controller |
| ci_enc_dim | Size of the hidden state in the controller encoder |
| s_min | the minimal event size for calcium events. |
-Not Frequently Changed Parameters:
| Name | Description |
|---|---|
| explore_method | The method used to explore hyper-parameters. Accepts two possible arguments - 'perturb' and 'resample' |
| explore_param | The parameters for the explore method defined by the 'explore_method' arg. When the 'explore_method' is set to 'perturb', the explore_param takes in a scalar between 0 and 1. A random number is then sampled uniformly from (1-explore_parama, 1+explore_param). This sampled number is then used as a factor which is multiplied to the HPs current value to perturb it. When the 'explore_method' is set to 'resample', the 'explore_param' must be set to None. In this condition, the new value of the HP is then obtained by resampling from a range of values defined for that parameter. |
| init_sample_mode | 'Rand','logrand' or 'grid', mode of preferred initialization. Or pass. |
| explorable | True or False, whether the 'explore' action should apply to this hyperparameter |
| limit_explore | If true, the new value of the HP after applying the explore method, cannot exceed the range defined by the "explore_range" arg. |
| explore_range | Uses tuple to indicate range, or list/nparray for allowable values |
| loss_scale | Scaling of loss |
| max_grad_norm | Maximum norm of gradient before gradient clipping is applied |
| cell_clip_value | Max value recurrent cell can take before being clipped |
-NeuroCAAS Parameters:
| Name | Description |
|---|---|
| Duration | Time allowed for completing a run. NeuroCAAS requires a preset computing duration to avoid unexpected long runs and save resources. RADICaL runs vary in the number of generations needed to converge. A run of RADICaL typically converges in 70 - 150 generations (approximately 3 - 6 hours). We recommend setting the “Duration” parameter to 6 hours (360 minutes). If a run is successfully finished, you can see the statement “Done.” printed in the logs and the pbt_best_model folder will be created in the results folder. If the actual run time is longer than the preset Duration, the run will not succeed, and the above results will not be present.The run will then need to be restarted with a larger Duration. |
| Dataset size | Immutable Analysis Environment storage space |
Other HPs which are set at their default values and cannot be changed by the user are described here
Troubleshooting
-Insufficient Instance Capacity: This error can occur due to insufficient instance capacity in the region. To resolve this issue, please wait for approximately 10 minutes and try starting the run again.
-Flat Outputs: If the inferred event rates are flat, the model is underfitting. Please try the following: (a) increase gen_dim to a higher number; (b) increase factors_dim to a higher number. Both (a) and (b) give the network more power to model complex dynamics; (c) increase num_workers. This facilitates hyperparameter search by increasing the number of single models trained simultaneously by PBT. (d) increase num_no_best_to_stop to a higher number, to give RADICaL more chances to overcome a local minima.
}
You must login to use an analysis.